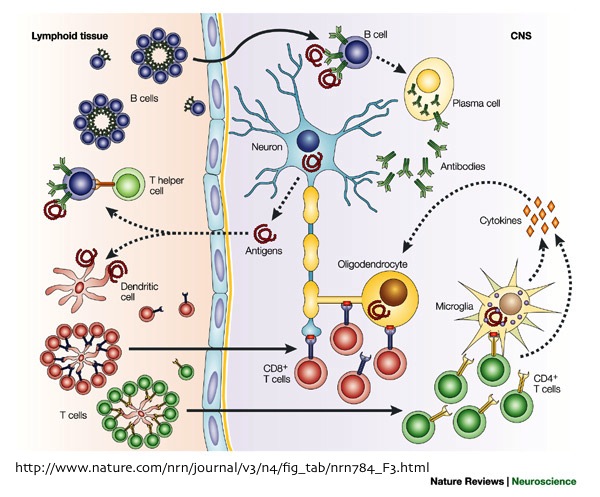
One thing that every immunologist agrees on is that immune cells are heterogeneous, and human peripheral blood mononuclear cells (PBMC) contain many different immune cell types. CD4+ T cells alone exist in dozens of uniquely functioning subsets. Receptor mediated signal transduction events in response to signals such as cytokines, chemokines, various receptor ligands, and engagement of the T cell or B cell receptors represent an important aspect of immune cell communication and the many types of immune cells present in human PBMC are differentiated by these responses. Trying to derive mechanisms from averages of complex PBMC populations obscures the understanding of the relationships that occur between expression of proteins and signaling states in unique cell populations and on the single cell level.
Dr. Gary Nolan and colleagues pioneered a method using flow cytometry to look at cell signaling in single cells using antibodies targeting phosphorylated protein sites, termed phospho-flow cytometry or “phosflow”. In phosflow, PBMCs or other cell populations are stimulated with signaling receptor ligands and/or antagonists for a period of time. Then the cells are fixed using paraformaldehyde-based buffers to lock the cells in their induced states of phosphorylation. The cells are then permeabilized and stained with fluorescently-labeled antibodies against the phosphorylated proteins, antigens to identify cell populations, and other proteins of interest, and analyzed on a flow cytometer.
Using this method, our lab has assessed the response to many cytokines by analyzing phosphorylation of the STAT family of transcription factors, as well as phosphorylation events that occur downstream from T cell receptor (TCR) and B cell receptor (BCR) engagement, and Toll-like receptor ligation, such as NF-kB (p65), ERK, p38, and ZAP70. BD Biosciences is a key resource for many of the reagents needed to perform these assays, providing several different buffers and antibodies against many important signaling targets.

There are several considerations for optimizing this protocol. The first is selecting the proper permeabilization method. Nolan and colleagues published a protocol in which 100% methanol is used for permeabilization following fixation, and this is the method I have used most frequently. One key advantage with this method is that following methanol permeabilization, the cells can be stored for an extended period of time at -20ºC or -80ºC prior to staining with the antibodies and running the flow cytometry. Thus, many samples can be stimulated on different days, but the flow cytometry analysis can be performed later in a large batch, in order to limit the variability between staining intensities that can otherwise occur between experiments. This is very important because when analyzing phosflow data, the fluorescence intensities which translate to magnitude of the phosphorylation of the protein target is often the key factor being analyzed, although the percentage of cells able to respond to a given signal can also be an important parameter for analysis. However, there are a number of commercially available buffer sets for these assays which should be selected based on the phospho-proteins and other antigens of interest.
Another important factor to consider when setting up these assays is selection of the target antigens for identifying cell populations of interest. Some antigens do not survive permeabilization with some of these reagents, including methanol. For instance, I have had poor luck trying to identify B cells using CD19 or monocytes using CD14. CD20 and CD33 have been used respectively as alternatives that work well with methanol.
Cytobank, an online resource dedicated to phosflow created by Gary Nolan’s lab, is great for assisting in selecting the proper antigens and antibody clones for phosflow. This website lists antibodies, including the phospho-target antibodies, that have been successfully and unsuccessfully tested with BD Biosciences’ various permeabilization buffers.
In summary, phosflow is an excellent method for analyzing signal transduction events in complex cell populations. Using single cell assays for studies on protein expression and signaling will drive our understanding of the immune system’s complexity to the next level.
Nolan lab Phosflow protocol:
Single-cell phospho-protein analysis by flow cytometry. Schulz KR, Danna EA, Krutzik PO, Nolan GP.Curr Protoc Immunol. 2012 Feb;Chapter 8:Unit 8.17.1-20.
BD Biosciences Phosflow Homepage:
http://www.bdbiosciences.com/research/phosflow/index.jsp?WT.ac=FP_Phosflow
Cytobank’s list of antibodies tested with various permeabilization buffers:
http://www.cytobank.org/facselect/
Further Reading:
Flow cytometric analysis of cell signaling proteins. Suni MA, Maino VC. Methods Mol Biol. 2011;717:155-69.
Single-cell, phosphoepitope-specific analysis demonstrates cell type- and pathway-specific dysregulation of Jak/STAT and MAPK signaling associated with in vivo human immunodeficiency virus type 1 infection. Lee AW, Sharp ER, O’Mahony A, Rosenberg MG, Israelski DM, Nolan GP, Nixon DF. J Virol. 2008 Apr;82(7):3702-12.







 Even though, iPSCs can be generated reproducibly, the efficiency of the reprograming remains low (i.e., around 1% using the original method). Furthermore, the initial generation of iPSCs utilized either retroviruses or lentiviruses, which both integrate into the host genome and might cause insertional mutagenesis, collectively creating a risk for translational applications. However, the beauty of the iPSC technology is its simplicity and reproducibility. Since 2007, iPSCs have been in the central focus of stem cell research and regenerative medicine, and researchers throughout the world have made a number of improvements to Yamanaka’s original protocol. Today, we have safer and more efficient methods for generation human iPSCs for disease modeling or cell replacement therapies, and by the look of how much has been achieved so far, the technology is only going to get better and more proficient. Considering that human embryonic stem cell derived approaches have been successfully used at the clinical level for cell replacement therapies, there is no doubt that iPSCs will eventually prove to be the ultimate source for regenerative medicine in the future.
Even though, iPSCs can be generated reproducibly, the efficiency of the reprograming remains low (i.e., around 1% using the original method). Furthermore, the initial generation of iPSCs utilized either retroviruses or lentiviruses, which both integrate into the host genome and might cause insertional mutagenesis, collectively creating a risk for translational applications. However, the beauty of the iPSC technology is its simplicity and reproducibility. Since 2007, iPSCs have been in the central focus of stem cell research and regenerative medicine, and researchers throughout the world have made a number of improvements to Yamanaka’s original protocol. Today, we have safer and more efficient methods for generation human iPSCs for disease modeling or cell replacement therapies, and by the look of how much has been achieved so far, the technology is only going to get better and more proficient. Considering that human embryonic stem cell derived approaches have been successfully used at the clinical level for cell replacement therapies, there is no doubt that iPSCs will eventually prove to be the ultimate source for regenerative medicine in the future.

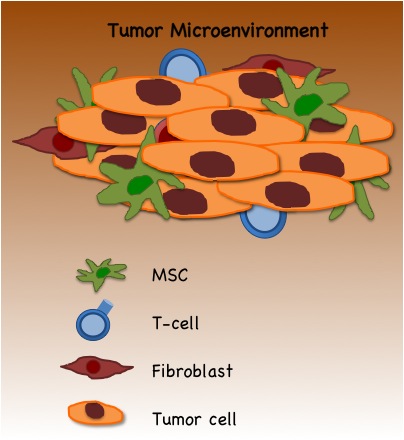
 The immune system is the body’s primary defender against cancer. In a recent article in Science magazine,
The immune system is the body’s primary defender against cancer. In a recent article in Science magazine,