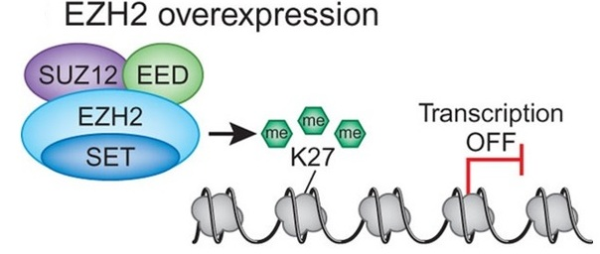
Although cancer is considered a disease of genetic defects, various studies have shown that epigenetic changes also play an important role in the onset and progression of cancer. Epigenetic modifications in mammals include DNA methylation and posttranslational histone modifications such as acetylation, methylation, phosphorylation, sumoylation, and ubiquitination. In human tumors, DNA methylation has been the most widely studied epigenetic modification. However, in recent years there has been a significant growth in our knowledge about the involvement of aberrant patterns of histone modifications in cancer development. In the nucleus, 147 base pairs of DNA are wrapped around an octamer of histone (H) proteins (two copies of each of H2A, H2B, H3 and H4) to form nucleosomes, which in turn are compacted further through several levels of higher-order packing (e.g., H1 aids formation of the 30-nm solenoid) to form chromatin. The accessibility of DNA within the nucleosome is in part controlled by the modifications of the histone proteins. Each histone protein in the nucleosome has a long “tail” that extends beyond the nucleosome. This lysine (K) rich amino-terminal “tail” undergoes various posttranslational modifications by acetyl groups, phosphate groups, and methyl groups. Among these various histone modifications specifically two modifications play crucial roles in the epigenetic control of cellular proliferation and differentiation. These include trimethylation of histone H3 lysine 27 (H3K27me3) which is catalyzed by the enhancer of zeste homolog 2 enzyme (EZH2), results in gene transcriptional repression; and methylation of histone H3 lysine 4 (H3H4me) which is catalyzed by the trithorax homolog myeloid-lymphoid leukemia (MLL), resulting in transcriptional activation. Several genes associated with development, stem cell maintenance, and differentiations are targets of H3K27 and H3K4 methylation. EZH2-mediated H3K27 methylation is also involved in X chromosome inactivation (Xi), a process in which one of the two X-chromosomes in the female cell is transcriptionally repressed, to generate transcriptionally inactive heterochromatin. In addition, as a catalytic subunit of epigenetic regulator Polycomb repressive complex 2, EZH2, a histone methyltransferase (an enzyme that transfers methyl groups), not only methylates histones H3 but also interacts with and recruits DNA methyltransferases to methylate CpG regions (C:cytosine, p:phosphpodiester bond, G:guanine) at certain EZH2 target genes and thereby causing transcriptional repression. Studies in cancer have indicated that deregulation of EZH2 contributes to a variety of tumor development and progression including breast, lung, prostate, pancreatic, and ovarian cancers as well as glioma, lymphoma, and sarcoma.
Overexpression of EZH2 was first reported in prostate an d breast cancer. In both cases increased expression was found to be associated with tumor invasiveness, metastasis, and poor clinical outcome. In addition, elevated expression of EZH2 is also reported in several other tumors including gastric, lung, bladder, and endometrial cancer. Gain of functions as a result of acquired mutations in EZH2 was reported in lymphoma and meyloid neoplasms. The best characterized mechanism by which EZH2 exerts its oncogenic function is by transcriptional repression of genes via its histone methyltransferase activity. Genes which get transcriptionally repressed by EZH2 include tumor suppressor genes ARF, p57KIP2, FBXO32, p27, and BRCA1. In addition, this enzyme also activates transcription of gene CCND1 (cyclin D1) driving cell-cycle progression.
d breast cancer. In both cases increased expression was found to be associated with tumor invasiveness, metastasis, and poor clinical outcome. In addition, elevated expression of EZH2 is also reported in several other tumors including gastric, lung, bladder, and endometrial cancer. Gain of functions as a result of acquired mutations in EZH2 was reported in lymphoma and meyloid neoplasms. The best characterized mechanism by which EZH2 exerts its oncogenic function is by transcriptional repression of genes via its histone methyltransferase activity. Genes which get transcriptionally repressed by EZH2 include tumor suppressor genes ARF, p57KIP2, FBXO32, p27, and BRCA1. In addition, this enzyme also activates transcription of gene CCND1 (cyclin D1) driving cell-cycle progression.
As oncogenic property of EZH2 is mediated through its enzymatic activity, inhibitors of EZH2 are currently under development targeting EZH2’s enzymatic activities. GSK126, a potent, highly selective small-molecule inhibitor of EZH2 methyltransferase activity exhibited promising response in diffuse large B-cell lymphoma (DLBCL) and DLBCL xenografts in mice. However, a recent study by Yan et al. (2013) demonstrated that the oncogenic function of EZH2 may not always be dependent on the enzymatic activities of EZH2. Therefore, therapeutic strategies targeting EZH2 should be designed based on its oncogenic activity via enzymatic properties as well as its function in transcriptional activation of genes involved in various oncogenic pathways.
References:
1. Ezhkova E, Pasolli HA, Parker JS, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. Vol. 136. United States; 2009:1122-1135.
2. Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. Vol. 42. United States; 2010:722-726.
3. Yan J, Ng SB, Tay JL, et al. EZH2 overexpression in natural killer /T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013.